Shaun Purcell - Genetics of Sleep Spindles
Genetics of Sleep Spindles
The NSRR is fortunate to have a diverse community of researchers, colleagues, and collaborators. A leading expert in his field, we were able to talk with Shaun Purcell about his research in the Genetics of Sleep Spindles. Thank you Shaun for your time and contributions!
Can you tell us a little about your group?
Shaun Purcell: My group develops statistical and computational tools for the design of genetic studies, the detection of gene variants influencing complex human traits and the dissection of these effects in the larger context of other genetic and environmental factors. I am currently an Associate Professor at the Icahn School of Medicine at Mount Sinai in New York and hold a joint appointment at Massachusetts General Hospital, Boston. Following an initial training in experimental psychology (University of Oxford and University College London), I've come to focus on neuropsychiatric genetics – previous work includes the development of the PLINK software package1 and studies of rare2–4 and common5 variation in schizophrenia, using genome-wide association study (GWAS) and exome sequencing approaches.
How is the NSRR helping your research?
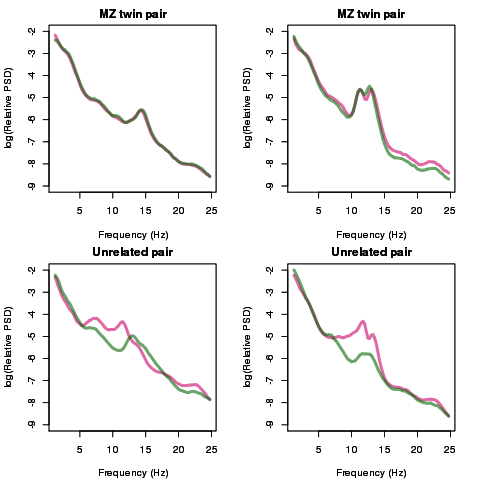
Shaun Purcell: With collaborators Dara Manoach, Robert Stickgold and others, we are currently leveraging National Sleep Research Resource data to investigate the genetic basis of brain activity during sleep. In particular, we are interested in sleep spindles – short bursts of 12-15Hz activity that characterize stage 2 non-REM sleep and can be detected from the electroencephalogram (EEG). Spindles appear to play important roles in memory and learning in healthy individuals; spindle abnormalities have also been implicated in neuropsychiatric diseases including schizophrenia. The extent and nature of spindle activity varies between people: for example, men show different profiles compared to women, and spindle activity generally declines with age. It is also increasingly clear that another major source of variability lies within our genomes. We aim to use NSRR data, combined with information from genetic studies, to identify the genes that drive variation in spindles. As previous work has shown, many aspects of sleep including spectral power and spindle activity are heritable phenotypes that can be reliably measured and so should be appropriate traits for further genetic analysis (Figure 1). By studying the shared genetic influences on sleep spindles and neuropsychiatric disease, we may be able to understand better the relationship between brain activity during sleep and disease risk.
What would you like to tell newcomers about the site?
Shaun Purcell: The NSRR website is a leading exemplar of how to share large and complex datasets among the research community. These data – collected across many years on thousands of individuals – have the potential to address many important scientific questions, and so it is great that they are being put to use. Our current research on the genetics of sleep spindles depends entirely on the NSRR – it simply would not be possible without it. Beyond just providing access to raw data, the NSRR really excels in making these data understandable and truly useable. It is easy to use, well documented and comprehensive, containing important meta-data about the protocols used in each study. I've been incredibly impressed by the active and ongoing efforts of the NSRR to help the research community to make the widest possible use of the data. The NSRR is a model for data sharing, and one that groups in other areas would do well to follow.
Figure 1. Spectral power during stage 2 non-REM sleep in identical (MZ) twins and unrelated individuals. The figure shows the spectral density for four pairs of individuals: the top two plots are for two MZ twin pairs, the lower two plots are for two age/sex-matched pairs of individuals. Lines indicate the relative activity across different frequency bands – from slow-wave sleep (0.5-1 Hz), to the spindle-related sigma-band (12-15Hz), to higher frequency beta oscillations (15Hz up to 25Hz as shown here). Each line represents one member of each pair: consistent with high heritability, whereas see considerable variation between unrelated individuals, MZ twins show strikingly similar profiles – the two twins in the top-left plot have effectively identical profiles. In our future work we hope to identify genes that drive these differences in spectral and spindle traits.
References
- Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455, 237–241 (2008).
- Purcell, S. M. et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190 (2014).
- Fromer, M. et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184 (2014).
- Purcell, S. M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Shaun Purcell